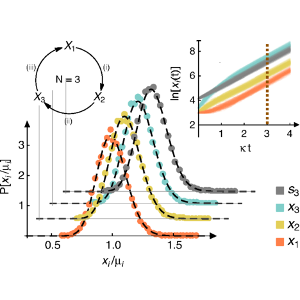
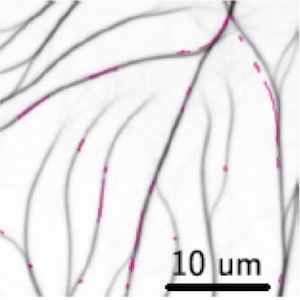
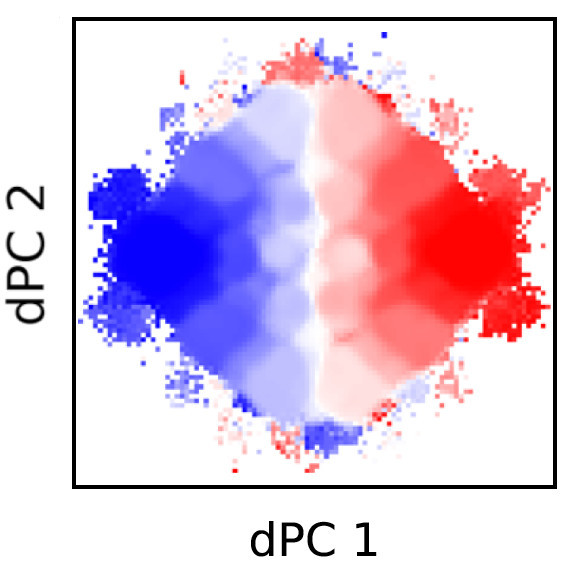
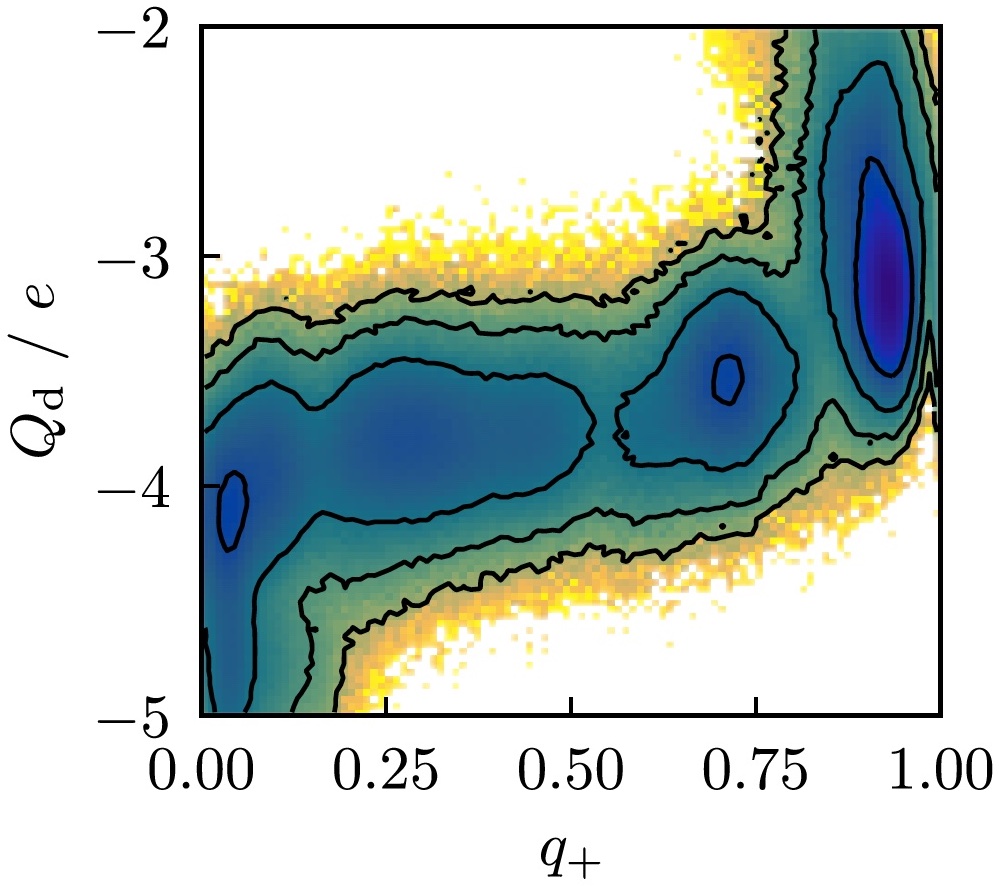
The Dinner group seeks to develop a theoretical understanding of how complex biological behavior arises from molecular interactions. Because the defining properties of living systems (growth, movement, and directed response to environmental stimuli) rely on irreversible energy consumption and dissipation, much of our research centers on stochastic processes far from equilibrium. We quantitatively analyze experimental data on living systems, construct physical models to interpret the observed statistics, and implement algorithms for efficiently simulating the dynamics of such models.
Research interests
Accelerating molecular simulations
We are developing, mathematically characterizing, and employing algorithms for enhancing the sampling of rare events and recovering their statistics.

People
Aaron Dinner
I am a Professor of Chemistry and Deputy Dean of the Physical Sciences Division at the University of Chicago. I also hold appointments in the James Franck Institute and the Institute for Biophysical Dynamics. My group’s most well known contributions are to machine learning methods for interpreting complex biomolecular simulations, sampling methods for systems far from equilibrium, and models of hematopoietic cell fate choice. Much of my current work is collaborative, and many of my students and postdoctoral scholars are jointly mentored by experimentalists and/or applied mathematicians. My honors include a Searle Scholarship, NSF CAREER Award, Sloan Fellowship, APS Fellowship, and participation in the 2010 and 2014 Latke-Hamantash Debates.
Prior to joining the faculty at Chicago, I obtained my undergraduate (AB in Biochemical Sciences, 1994) and graduate (PhD in Biophysics, 1999) degrees at Harvard University, where I worked with Martin Karplus on Monte Carlo methods and their application to protein folding. Subsequently, I pursued postdoctoral studies at the University of Oxford (1999-2001), where I used hybrid quantum-mechanical/molecular-mechanical (QM/MM) methods to elucidate mechanisms of DNA repair, and the University of California, Berkeley (2001-2003), where I worked with David Chandler on transition path sampling and Arup Chakraborty on models of T lymphocyte signaling.
professor | since 2003
Ian Bongalonta
I am developing computational methods to interpret 2D infrared spectra and applying them to studying intrinsically disordered proteins.
graduate student | since 2023Chris Chi
I am developing and applying methods for sampling nonlinear dynamical systems consistent with contraints, including experimental data.
graduate student | since 2019Carlos Floyd
I am using theory and simulation to understand hallmark features of active biological systems, including force generation and learning.
postdoctoral scholar | since 2021Noah Gamble
I employ genomics, phenomenological modeling, and biochemical approaches to investigate gene expression and its consequences for cellular plasticity in the immune system.
graduate student | since 2020Spencer Guo
I am studying voltage-sensing with machine learning methods for estimating kinetics and elucidating mechanisms from short-trajectory data.
graduate student | since 2021Qi-Nan Huang
I am using machine learning to analyze libraries of protein-protein interactions.
graduate student | since 2025Kwanghoon Jeong
I am studying calcium-binding proteins with machine learning and molecular dynamics simulations.
graduate student | since 2022Darren Liu
I am combining experiments and simulations to study the physics of directed evolution.
graduate student | since 2023Chatipat Lorpaiboon
I use machine learning and transition path theory to develop data-driven methods for analyzing stochastic dynamics.
graduate student | since 2018Ondrej Maxian
I am developing models for cytoskeletal dynamics and fitting them to experimental data with Markov chain Monte Carlo methods.
postdoctoral scholar | since 2024Nhan Nguyen
I am studying voltage-gated ion channels with machine learning and molecular dynamics simulations.
graduate student | since 2024Zihan Pengmei
I am exploring neural-network architectures for learning protein dynamics.
graduate student | since 2023Nico Romeo
I am developing machine learning methods for characterizing nonlinear dynamics.
postdoctoral scholar | since 2024Sarah Root
I am defining inflammatory cell states with microscopy and machine learning.
graduate student | since 2023Jordan Shivers
I use theoretical and computational tools from soft matter physics, nonequilibrium thermodynamics, and machine learning to study emergent behavior in active assemblies of cellular components.
postdoctoral scholar | since 2022Nils Strand
I am developing tensor-network methods for density estimation to address biological problems.
postdoctoral scholar | since 2023Weizhou Wang
I am developing generative machine learning methods for path sampling.
graduate student | since 2024Yihang Wang
I am developing generative machine learning methods for protein ensembles.
postdoctoral scholar | since 2022
Recent publications
Selected recent publications are highlighted below. For a full list, see PubMed or Google Scholar.
Inexact iterative numerical linear algebra for neural network-based spectral estimation and rare-event prediction
John Strahan, Spencer C Guo, Chatipat Lorpaiboon, Aaron R Dinner, and Jonathan Weare
Journal of Chemical Physics 159, 014110 (2023) Link
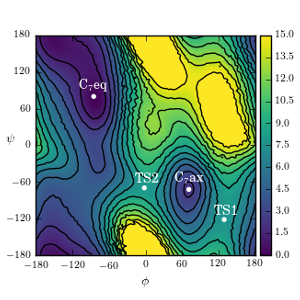
Dynamics of activation in the voltage-sensing domain of Ci-VSP
Spencer C Guo, Rong Shen, Benoit Roux, and Aaron R Dinner
Nature Communications 15, 1408 (2024) Link
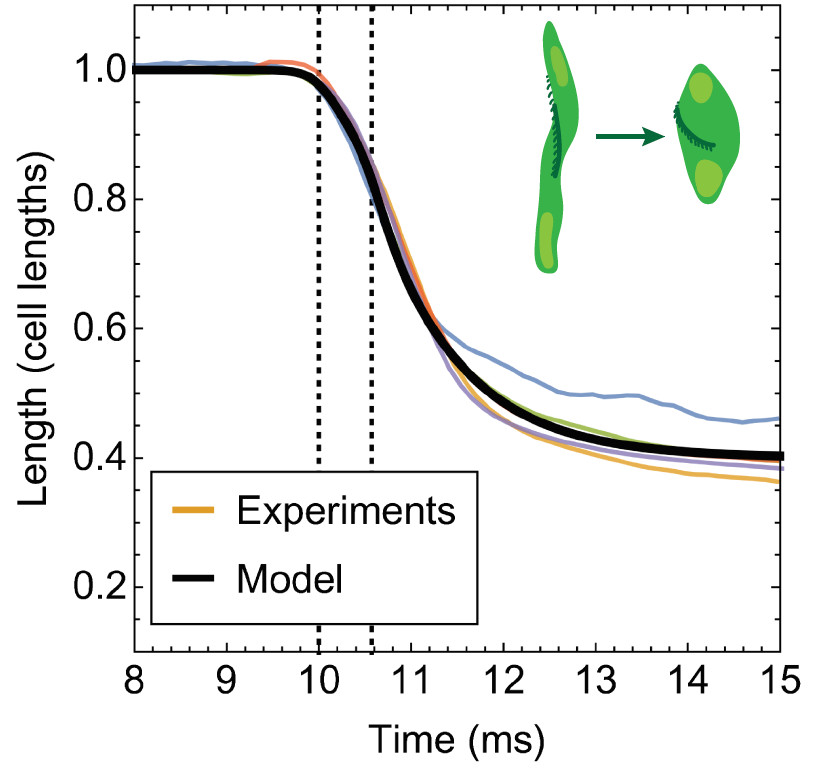
A unified model for the dynamics of ATP-independent ultrafast contraction
Carlos Floyd, Arthur T Molines, Xiangting Lei, Jerry E Honts, Fred Chang, Mary Willard Elting, Suriyanarayanan Vaikuntanathan, Aaron R Dinner, and Saad Bhamla
PNAS 120, e2217737120 (2023) Link
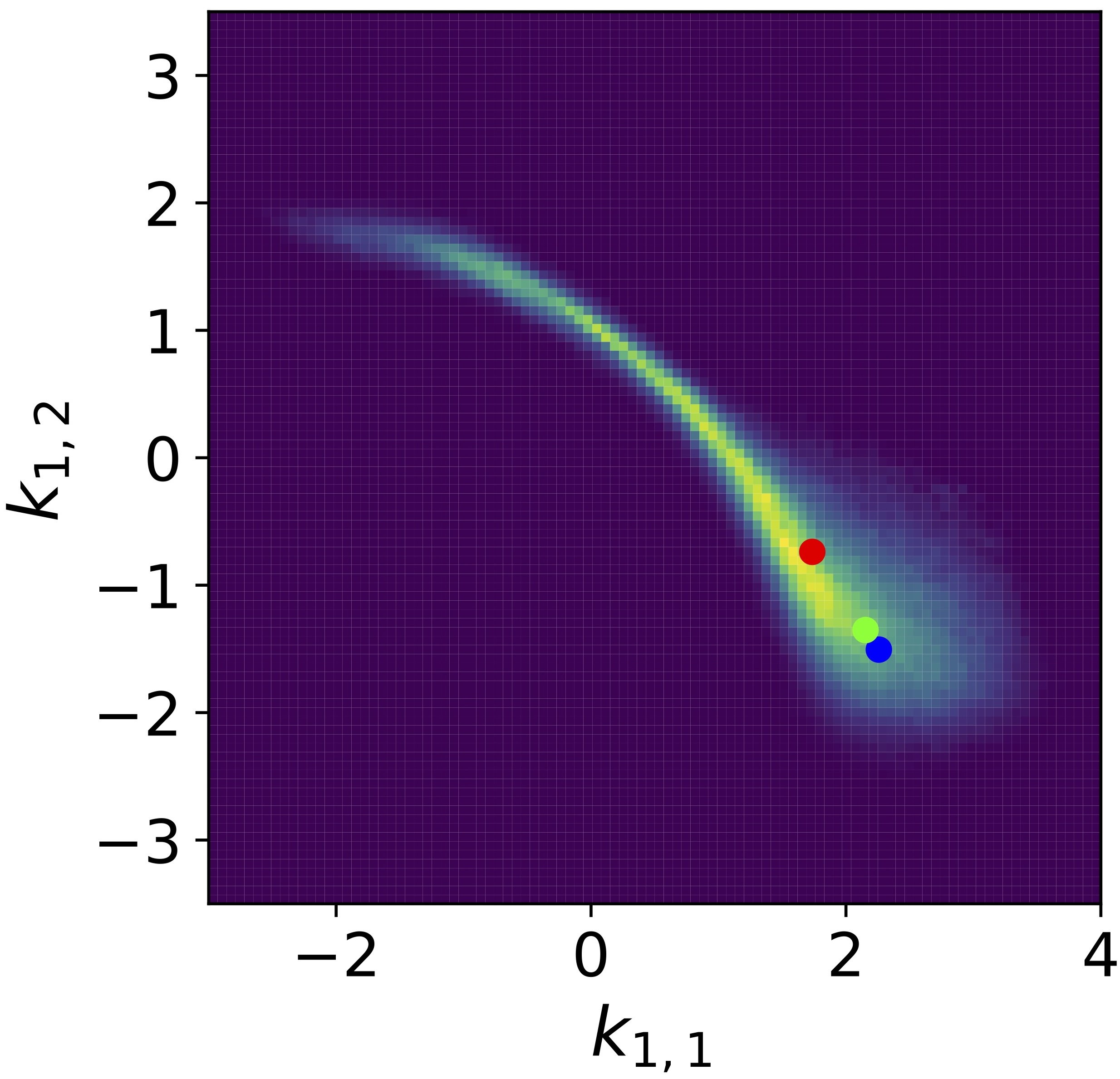
Sampling parameters of ordinary differential equations with Langevin dynamics that satisfy constraints
Chris Chi, Jonathan Weare, and Aaron R Dinner
arxiv.org (2024) Link